¶ Regions-of-Interest (ROIs)
EMBED contains a number of ROIs that were generated when the annotating radiologist made an indication during their exam. This page discusses what they represent, how they were derived, and how to use them.
¶ What do they represent?
EMBED ROIs were collected at the time of screening so they represent a snapshot of the radiologist's decision making in that moment. Due to this, they're most common in (but not exclusive to) screening exams with BI-RADS A findings.
¶ How were they derived?
All ROIs in EMBED were mapped from elliptical coordinates to rectangles and were derived from one of two possible source objects:
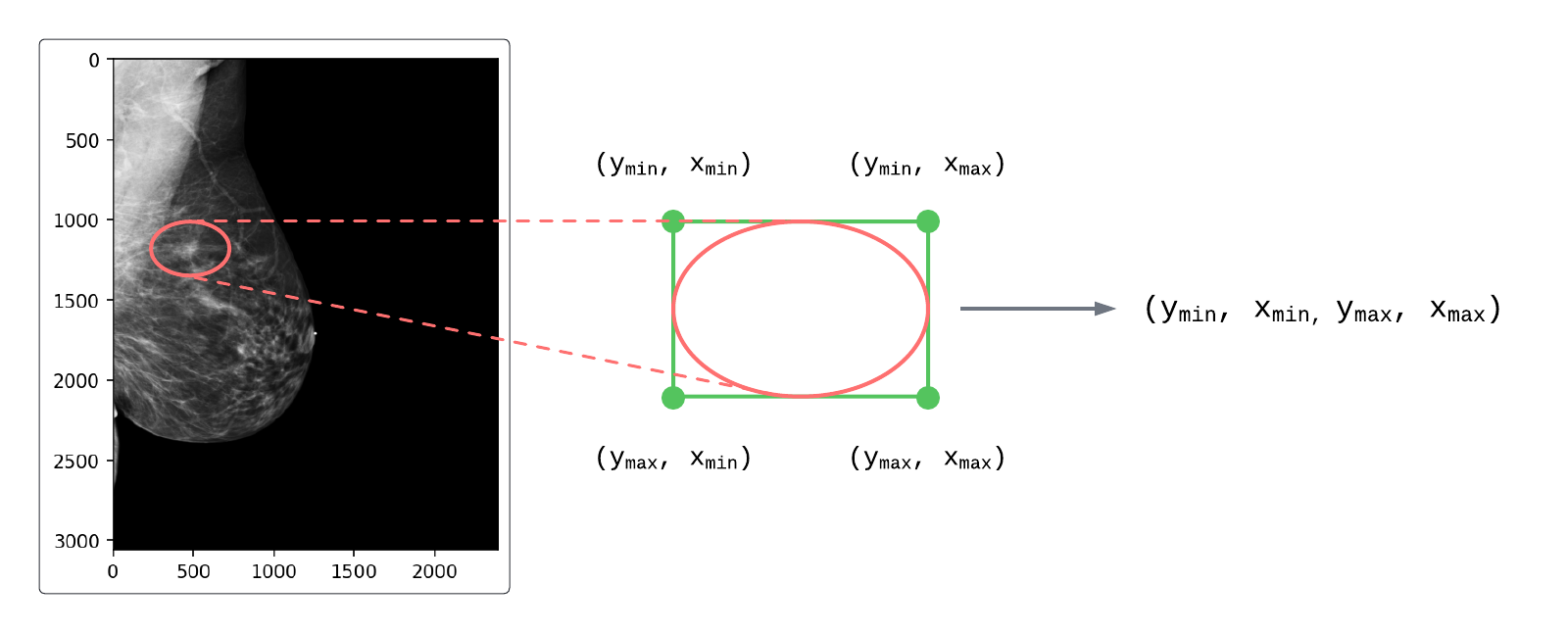
Figure 1: Mapping elliptical ROI coordinates to rectangular coordinates
- Secondary Screen Capture (SSC) Images
Some ROIs were derived from SSC images. These were generated when an ROI was created and saved as a separate image (including the ROI baked into the pixel data) in the PACS. These images had exam-level identifiers present, but did not have a direct match to their source DICOMs in their metadata. To handle this, structure similarity index measure (SSIM) was used to match SSCs to their source DICOMs (example shown in Figure 2).
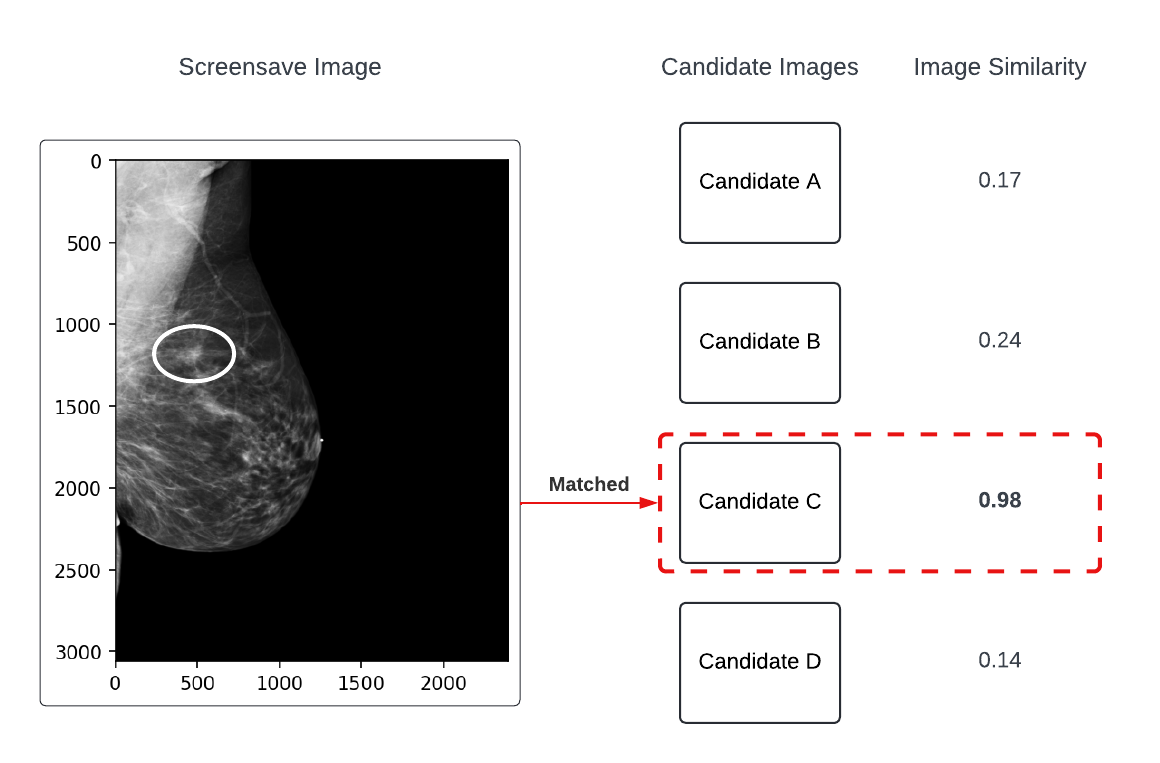
Figure 2: Example of the SSC-to-DICOM matching process
- DICOM Annotation Objects
Some ROIs were derived from DICOM annotation objects which contained no pixel data and instead had a linked SOPInstanceUID and a set of elliptical coordinates. These coordinates were mapped from ellipses to rectangles (as shown in Figure 1) and matched back to their source DICOMs by SOPInstanceUID.
¶ Usage
Each ROI is given in the form:
[ymin, xmin, ymax, xmax]
And individual ROIs are stored as nested lists within a container list:
[[ROI_1], [ROI_2], ...]
These coordinates are stored as strings in the ROI_coords column. In Python, these strings can easily be parsed into nested lists using the ast.literal_eval() function (link to documentation) from the built-in ast package.

Figure 3: Example of ROI coordinate nesting
¶ Plotting ROIs on Source Images
The following code snippet shows an example of how ROIs can be plotted on the source images to inspect them. Here, we use a Rectangle (link to documentation) object from matplotlib.patches as an easy way to outline the ROIs. Figure 4 below the snippet has example outputs from this function.
Code Snippet: Example function to visualize an ROI on an image
from matplotlib.patches import Rectangle
def plot_rois(row: pd.Series) -> None:
# extract pixel array from the DICOM
image: np.ndarray = pydicom.dcmread(row.anon_dicom_path).pixel_array
# extract ROI coordinates as a list of integers
roi_list: list[list[int]] = ast.literal_eval(row.ROI_coords)
# create a figure and plot the image
fig, ax = plt.subplots(1, 1, dpi=130)
ax.imshow(image, cmap='gray')
# disable axis ticks on plot
ax.set_xticks([])
ax.set_yticks([])
# zip the rois and their match levels together
for roi in roi_list:
# unpack ROI values
ymin, xmin, ymax, xmax = roi
# format the roi into a patch
roi_patch = Rectangle(
(xmax, ymax),
xmin - xmax,
ymin - ymax,
edgecolor='xkcd:bright red',
fc='None',
label='ROI'
)
# add the patch to the axes
ax.add_patch(roi_patch)
ax.legend()
fig.show()
plot_rois(row)
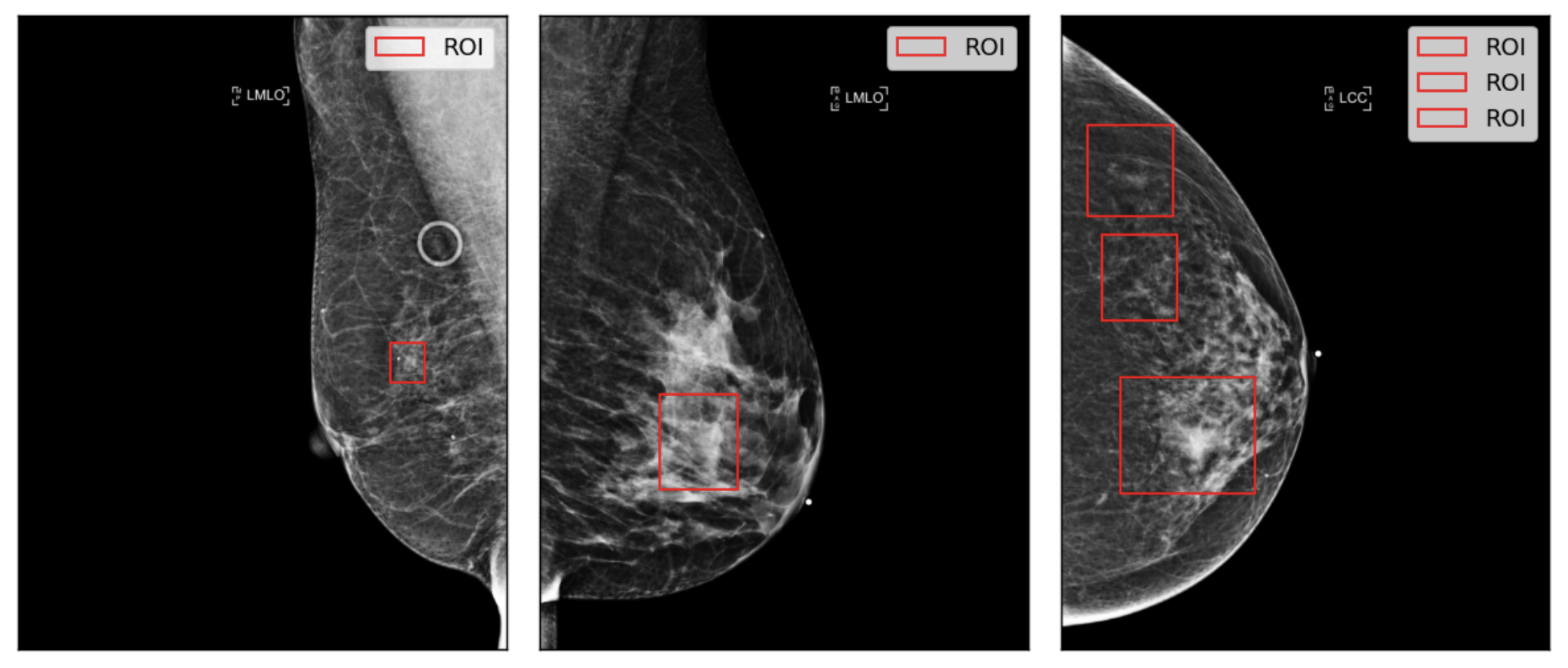
Figure 4: Example of ROI visualizations on source images
¶ Extract ROIs as Patches
One way to work with ROIs is to extract the pixels within the ROI and saving them to a new image (often called a 'patch').
Code Snippet: Visualizing ROIs as image patches
# parse ROI coordinates
roi_coords: list[list[int]] = ast.literal_eval(row.ROI_coords)
# load full image from the DICOM pixel array
image: np.ndarray = pydicom.dcmread(row.anon_dicom_coords).pixel_array
# iterate over nested ROIs
for roi in roi_coords:
# unpack ROI points
y_min, x_min, y_max, x_max = roi
# use array indexing to extract the image within the ROI
patch = image[y_min:y_max, x_min:x_max]
# plot the patch
plt.imshow(patch, cmap="gray")
plt.show()
Since ROIs can have varying dimensions, they are commonly tissue- or black-padded to normalize their dimensions. Figure 5 shows some examples of these different approaches and the snippets below contain example code for how this could be done.
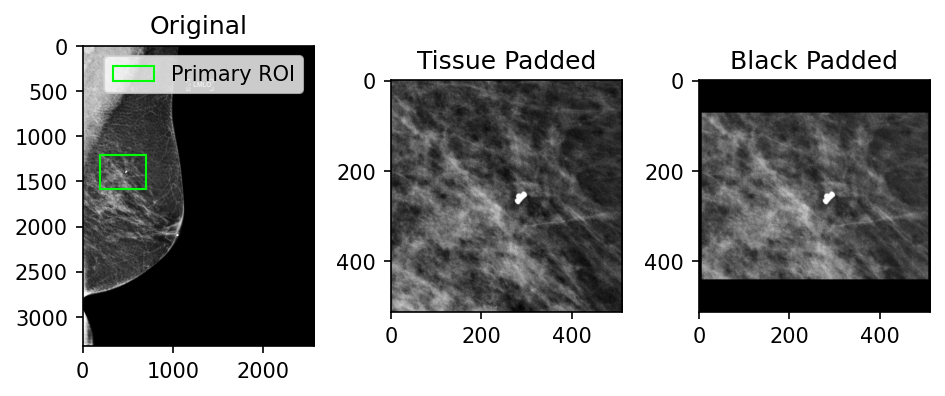
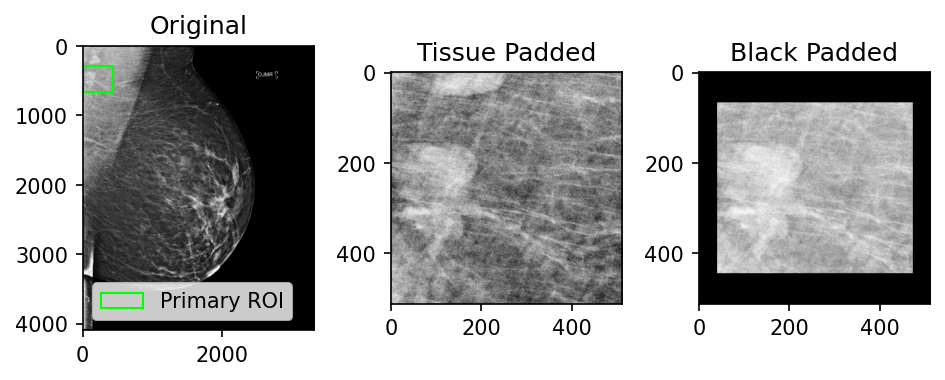
Figure 5: Examples of patch tissue-padding versus black-padding (512x512 patch size)
Code Snippet: Example function to tissue-pad a patch
def tissue_pad_patch(img: np.ndarray, roi: list[int], patch_size: int) -> np.ndarray:
"""
function to tissue pad a patch given an image, a single ROI in the
form [y_min, x_min, y_max, x_max] and the desired patch size
"""
# unpack roi
y_min, x_min, y_max, x_max = roi
# get the center y, x
y_center: int = (y_min + y_max) // 2
x_center: int = (x_min + x_max) // 2
# get initial patch coords
half_size: int = patch_size // 2
patch_y_min: int = y_center - half_size
patch_x_min: int = x_center - half_size
patch_y_max: int = y_center + half_size
patch_x_max: int = x_center + half_size
# correct the patch coords to make sure they fall within the image
patch_y_max: int = max(min(patch_y_max, img.shape[0]), patch_size)
patch_x_max: int = max(min(patch_x_max, img.shape[1]), patch_size)
# ensure y_min is less than y_max - min_roi_size
# and greater than 0
patch_y_min: int = max(min(patch_y_min, patch_y_max - patch_size), 0)
patch_x_min: int = max(min(patch_x_min, patch_x_max - patch_size), 0)
return img[patch_y_min:patch_y_max, patch_x_min:patch_x_max]
Code Snippet: Example function to black-pad a patch
def black_pad_patch(img: np.ndarray, roi: list[int], patch_size: int) -> np.ndarray:
"""
function to black pad a patch given an image, a single ROI in the
form [y_min, x_min, y_max, x_max] and the desired patch size
"""
# unpack roi
y_min, x_min, y_max, x_max = roi
# initialize our patch as an empty array of zeros
patch: np.ndarray = np.zeros((patch_size, patch_size), dtype=np.uint8)
half_size: int = patch_size // 2
# we need to handle cases where the ROI is larger than the patch size
# check if the height of the roi exceeds the patch size
if (y_max - y_min) > patch_size:
# get the center coordinate
y_center: int = (y_max + y_min) // 2
# clip to the roi size
y_min: int = y_center - half_size
y_max: int = y_center + half_size
# check if the width of the roi exceeds the patch size
if (x_max - x_min) > patch_size:
# get the center coordinate
x_center: int = int((x_max + x_min) / 2)
# clip to the roi size
x_min: int = x_center - half_size
x_max: int = x_center + half_size
# this gives us the offset between the patch edge and the roi so we can center it
patch_height: int = y_max - y_min
patch_width: int = x_max - x_min
half_y_diff: int = (patch_size - patch_height) // 2
half_x_diff: int = (patch_size - patch_width) // 2
# extract the roi from our original image and center it on the black patch array
patch_contents: np.ndarray = img[y_min:y_max, x_min:x_max]
patch[
half_y_diff:half_y_diff + patch_height,
half_x_diff:half_x_diff + patch_width,
] = patch_contents
return patch